Oncogenetics and forensic genetics group
Our research group is mostly focused on the genetic and epigenetic biomarkers with two main applications – oncology and forensics. As tumorigenesis is started by genetic mutations, diagnostic, and curative part of oncology relies on results from genetic testing of samples from tissue biopsy or liquid biopsy of tumour. The amount of information that can be gained from biopsy increased exponentially by introducing massively parallel sequencing into the routine clinical practice. However, there is still room for improvement because of the inherent characteristics of tumours. Quantity and quality of attainable nucleic acid pose technical limitations and many tumours do not have biomarkers that would make their treatment sufficiently efficient. Thus, we are seeking for new genetic and epigenetic biomarkers and we are striving for better analytical and procedural parameters of genetic methods.
Apart from oncology, we apply genetic and epigenetic methodology to crime scene trace characterisation in forensics. Epigenetics is study of heritable phenotype changes that are not governed by changes in the sequence of nucleotides in DNA. It looks at gene regulation, at changes of phenotype without changes in genotype. By metaphor, genetics provides sheet music but epigenetics decides how to play it. Important thing is that epigenetics is influenced by environmental and lifestyle factors and at least partially reversible. Despite no match with suspect profile or criminalistic database, DNA can still be useful as an intelligence tool. By epigenetic testing we are trying to find information that would help investigation, such as characterisation of perpetrator or crime circumstances.
- Nucleic acids
- Oncological and forensic biomarkers
- Genotyping
- Massively parallel sequencing
- Bayesian interpretation
Massively parallel sequencing of oncomarkers
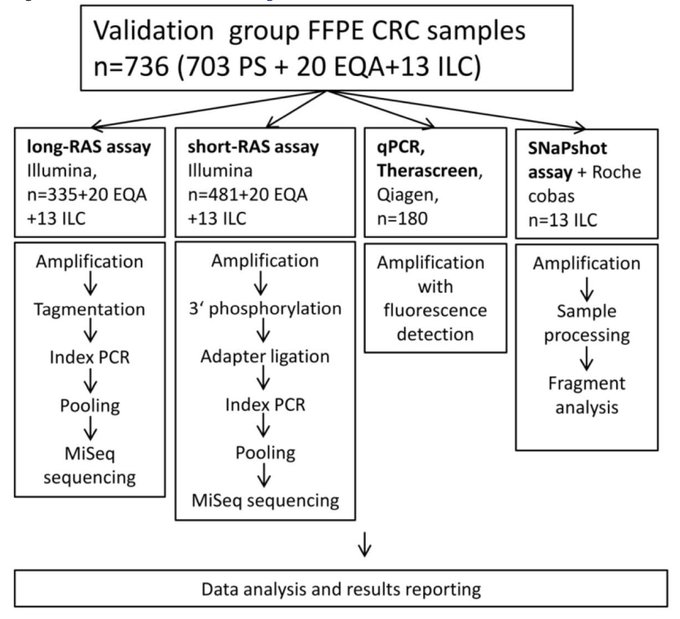
Companies currently offer massively parallel sequencing (MPS) kits, applicable to whole genome, exome, or subset of genes - gene panels. Such kits are designed for international clients what sometimes does not reflect the requirement of clinicians, as expressed by Blue Book of the Czech Oncological Society and healthcare insurance agreement. Also, their genotyping success rate does not reach the numbers that we routinely attained by qPCR methods applied to single variant testing from tumour sample. Thus, we are trying to modify the preanalytical and analytical steps of the methods, such as DNA extraction, library preparation, and bioinformatics pipelines, to allow gaining information from challenging samples, such as liquid biopsy. Also, by application of MPS to well characterised clinical cohorts, we are trying to find new biomarkers for biological treatment of tumour.
Droplet digital PCR of oncomarkers
Droplet Digital PCR technology (ddPCR) is PCR method that utilizes a water-oil emulsion droplet microfluidic system as reagents reservoir. Sample (DNA) and reagents are divided into thousands of nanoliter-sized droplets and PCR amplification is carried out within each droplet. The advantage is 1) absolute quantification of target DNA copies per input sample without the need for calibration standards and 2) lower limit of detection of the target compared to qPCR.
ddPCR technology will be used for detection of rare specific mutations from liquid biopsy (e. g. for patients suffered from lung cancer with resistant mutations in EGFR gene caused by treatment with tyrosine kinase inhibitors). The release of the result will be faster and with higher sensitivity, liquid biopsy less invasive for the patient, both important for following patient treatment.
Hematopoietic stem cells mutations, aging, and cancer
Aging represents the single biggest risk factor for chronic diseases like cancer. While every cell have the carcenogenic potential, clonal dominance of expanded mutant stem and progenitor cell population occuring in aging tissues may be an early event in the development of hematologic cancers. Such clonal dominance may be present years before cancers become clinically important, and is gradually manifested by genesis of genome desintegration and impaired organ maintenance.
Negative consequences of aging arise from interplay of genetics, epigenetics, microenvironment, and immune system. Better understanding of the interaction between cancer stem cells (tumor initiating cells) and tumor immunology may help to eradicate the minor subpopulations that escape conventional therapy and cause drug resistance and metastasis.
We want to test by massively parallel panel sequencing the blood and atherosclerotic plaques samples of patients with carotid atherosclerosis, stroke, and controls for clonal hematopoiesis of indeterminate potential.
Epigenetic biomarkers of age
Epigenetic age prediction from biological trace on crime scene can narrow the pool of potential suspects, reducing thus the effort in the lab in terms of money and time, and reducing preanalytical effort in terms of buccal swab sampling. Age prediction relies on fact that activity of certain genes changes during the lifetime, based on increase or decrease of methylation of cytosines (CpGs) in promotors of genes. Epigenetic clocks based on microarray DNA testing can predict age with high precision. However, such analysis of dozens of CpGs is inapplicable for forensic practise, where low quality and quantity of DNA is the main limiting factor. Thus, we are currently trying to find a small set of CpG with high predictive power that can be analysed by a sample-sparing method, massively parallel sequencing from blood and sperm traces.
| Project: | Genetic and epigenetic biomarkers in health and disease |
|---|---|
| Supervisors: | Slavkovský Rastislav Ph.D., Džubák Petr M.D., Ph.D., Hajdúch Marián M.D., Ph.D., Drábek Jiří Ph.D., Koudeláková Vladimíra Ph.D. |
| Available: | 5 |
| Intended for: | Doctoral training |
| Project: | Genetic and epigenetic biomarkers of cancer diseases |
|---|---|
| Supervisors: | Drábek Jiří Ph.D. |
| Available: | 1 |
| Intended for: | Doctoral training |
| Summary: | 1 place in full-time study |
| Project: | Genetic and epigenetic biomarkers of cancer diseases |
|---|---|
| Supervisors: | Slavkovský Rastislav Ph.D., Džubák Petr M.D., Ph.D., Hajdúch Marián M.D., Ph.D. |
| Available: | 3 |
| Intended for: | Doctoral training |
| Summary: | 3 places in full-time study |
| Project: | Epigenetic biomarkers |
|---|---|
| Supervisors: | Drábek Jiří Ph.D. |
| Available: | 1 |
| Intended for: | Master training |
| Project: | Epigenetic biomarkers |
|---|---|
| Supervisors: | Drábek Jiří Ph.D. |
| Available: | 1 |
| Intended for: | Master training |
| Project: | Genetic biomarkers |
|---|---|
| Supervisors: | Drábek Jiří Ph.D. |
| Available: | 1 |
| Intended for: | Master training |
| Project: | Genetic biomarkers |
|---|---|
| Supervisors: | Drábek Jiří Ph.D. |
| Available: | 1 |
| Intended for: | Master training |
| Project: | Genetic and epigenetic biomarkers in cancer |
|---|---|
| Supervisors: | Drábek Jiří Ph.D., Slavkovský Rastislav Ph.D., Hajdúch Marián M.D., Ph.D. |
| Available: | 3 |
| Intended for: | Doctoral training |
| Summary: | Clonal hematopoiesis of indeterminate potential (CHIP) has recently been described as a common phenomenon associated with aging. It is characterized by the accumulation of somatic mutations in cells of the hematopoietic system. Although CHIP is manifested by the expansion of certain cell clones, this condition is not accompanied by any morphological features of hematological neoplasia. However, it has been shown that the incidence of clonal hematopoiesis correlates with increased overall mortality and the risk of developing malignant transformation of hematopoietic cells as well as cardiovascular disease, such as ischemic stroke. To what extent and by what mechanisms clonal hematopoiesis contributes to disease development remains a question of current research. The main aim of the project will be to pinpoint the principal cells carrying CHIP somatic mutations, and to study their role in development and maintenance of atherosclerotic plaques, especially of those involved in development of stroke. The comparison of the phenotype of CHIP positive and negative cells will be of special interest. The use of cellular models not only include different types of leucocytes but circulatory progenitor endothelial cells as well. The study will involve elderly subjects with the positive presence of CHIP (>65 years). Subject will be characterized based on the presence or absence of ischemic stroke and the presence or absence of carotid stenosis by our clinical collaborators. The presence of somatic variants in 38 selected genes associated with CHIP will be tested in subjects of interest within our research group. The project will use various techiques including FACS, MACS, cell cultures, DNA isolation from small amount of cells, a highly sensitive sequencing method for DNA genotyping allowing detection of variant with less than 1% allelic frequency, DNA/RNA sequencing library preparation, deep massively parallel sequencing of panel of genes using unique molecular barcodes/indices, RNAseq, bioinformatics and data analysis with possibilities of calculations using high performance computing cluster, data management and statistical evaluation. |